Studying the Fungi kingdom is important, because they have so many different ecological roles, including decomposers, symbiotes and parasites. There are also more than 1 million different species of fungi, so researchers need to have high-throughput methods to explore this diversity [1]. One such method is next-generation sequencing.
In this blog, we’ll go over why and how researchers sequence for fungi, what the ITS and 18S genes are, how to choose between them and how Genohub can help with your fungal sequencing project.
Why perform sequencing for fungal community analysis?
Fungal sequencing can be used to discover novel fungal species, quantify known fungi, explore the structure of fungal communities, and determine the roles of fungi in nature. In addition, it’s important to study these communities for human health, as there are some fungi that are resistant to antifungal drugs and others that are involved in plant diseases [2]. Thus, sequencing for fungi is relevant for multiple fields, including environmental conservation, agriculture, and microbiology.
Both ITS and 18S sequencing are well-established methods for studying fungal communities, as focusing on these genes is a simple way to identify fungi within complex microbiomes or environments that would otherwise be difficult to study [3]. For example, this type of specific amplicon sequencing enables the analysis of the fungal community within very mixed environmental samples, such as soil or water.
What are ITS and 18S?
The internal transcribed spacer (ITS) region and the 18S ribosomal RNA gene are used as biomarkers to classify fungi.
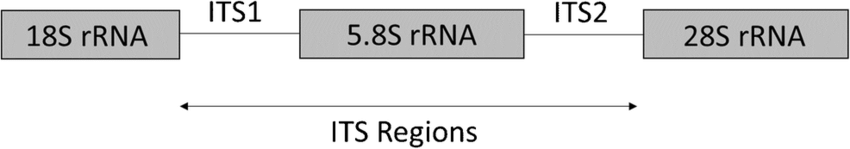
As seen in Figure 1, the ITS region includes ITS1 and ITS2, the spacer genes located between the small-subunit rRNA and large-subunit rRNA. Generally, the ITS1/ITS4 primers are used for amplification of the ITS region, although they can be substituted with the universal primers ITS2, ITS3, and ITS5 [4].
The 18S ribosomal RNA (18S rRNA) gene codes for a component of the small 40S eukaryotic ribosomal subunit and has both conserved and variable regions. The conserved regions can reveal the family relationship among species, whereas the variable regions will show the disparities in their sequences. Regarding the variable regions, 18S rRNA gene has a total of nine, V1-V9. The regions V2, V4 and V9 together are useful for identifying samples at both the family and order levels, while V9 seems to have a higher resolution at the genus level [5].
How to choose between ITS and 18S?
Although both ITS and 18S rRNA have proven useful for assessing fungal diversity in environmental samples, there are enough differences between them that researchers may choose to focus on only one, although sequencing for both is an option as well.
There was relatively low evolutionary pressure for the ITS1 and ITS2 sequences to remain conserved, so the ITS region tends to be hypervariable between fungal species while remaining moderately unchanged among individuals from the same species. It is therefore very well suited as a marker for species identification in the classification of fungus and is often used to study relative abundance of fungi as well [2]. This can be useful if you need to perform a survey for genetic diversity at the species level or even within a species.
On the other hand, there was significant evolutionary pressure for the 18S rRNA gene to remain highly conserved as a component of the small eukaryotic 40S ribosomal subunit, an essential part of all eukaryotic cells. Due to this pressure, 18S is considered a potential biomarker for fungi classification above the species level and is often used in wide phylogenetic analyses and environmental biodiversity screenings [5].
In summary, the ITS region is mainly used for fungal diversity studies, while 18S rRNA is mainly used for high resolution taxonomic studies of fungi.
How can Genohub help?
Genohub’s ITS and 18S sequencing partners are experts in every step of the amplicon sequencing process, including extraction, PCR amplification and library preparation using validated primers based on the literature, and data analysis, including taxonomic assignment, diversity and richness analysis, comparative analysis, and evolutionary analysis. Our partners have experience extracting from many different types of environmental and biological samples, including soil, water, sludge, feces, and plant and animal tissue, but they can work just as well with DNA samples that you extract yourself.
We know that each research project is unique, so we have partners who are also open to working with your custom primers or your custom analysis needs! Get started today by letting us know about your ITS or 18S sequencing project here: https://genohub.com/ngs/ .