Illumina sequencing is by far the most common next-generation sequencing technique used today, as it extremely accurate and allows for massively parallel data generation, which is incredibly important when you’re working with a dataset the size of a human genome!
That said, there are inherent shortcomings that exist in the typical Illumina sequencing workflow. Illumina uses a very high number of short sequencing reads (usually about 150 bp for whole genome sequencing) that are then assembled together to cover the entirety of the genome. The fact that traditional Illumina can’t be used to identify long-range interactions can cause issues in some cases, such as samples with large structural variants or in phasing haplotypes.
However, a revolutionary new library preparation method designed by 10x Genomics can effectively solve these types of issues. The 10X Genomics GemCode technology is a unique reagent delivery system that allows for long-range information to be gathered from short-read sequencing. It does through usage of a high efficiency microfluidic device which releases gel beads containing unique barcodes and enzymes for library preparation. It then takes high molecular weight DNA, and partitions it into segments that are about 1M bp long; from here, these segments of DNA are combined with the gel beads. This means that each read that comes from that segment of DNA has its own unique barcode, which gives us knowledge about long-range interactions from traditional short reads.
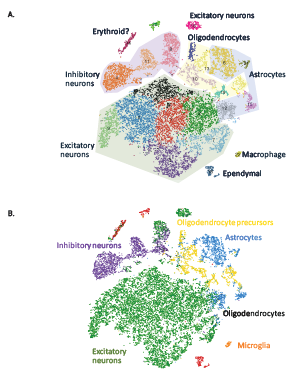
Figure 1: Projections of 20,000 brain cells where each cell is represented as a dot. (A) Shading highlights major clusters identified in the dataset. (B) Cells were colored based on their best match to the average expression profile of reference transcriptomes [2].
Traditional Illumina sequencing will still likely reign supreme for run-of-the-mill applications, since at this point it is still more cost effective. However, the 10x system is gaining in popularity for specialized applications where understanding structural variants or single cell sequencing is important to the goals of the project. We’ve certainly noticed an uptick in projects that require 10x technology recently, and we look forward to seeing the advances that can be made with this amazing technology.
If you’re interested in projects using 10x Genomics sequencing tech, please contact us at projects@genohub.com for more information!
Hi Genohub Blog, Can you be so kind and let me know if your company renders service of TCR repertoire profiling? We will need to quantify the number of T cells carrying a specific Vbetta chain in 40 samples of 7ml of human blood and 40 samples of 3mm>3 formalin fixed/ paraffin embedded human biopsies.
Thank you for your answer.
Kind regards,
Adam Friedmann
Professor of Genetics & Virology
Scientific advisor
*NeoTX* 2 Pekeris Street Rehovot, Israel 7670202
Phone: +972 3 912 5853
Fax: +972 8 936 8584
My phone +972 52 5761313
LikeLike